Note
Click here to download the full example code
Comparisons of decision functions¶
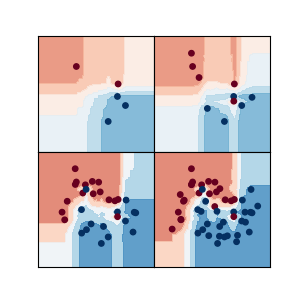
This example allows to compare the decision functions of several random forest types of estimators. The following classifiers are used:
- AMF stands for AMFClassifier from onelearn
- MF stands for MondrianForestClassifier from scikit-garden
- RF stands for RandomForestClassifier from scikit-learn
- ET stands for ExtraTreesClassifier from scikit-learn

import sys
import numpy as np
import matplotlib.pyplot as plt
import logging
from sklearn.preprocessing import MinMaxScaler
from sklearn.ensemble import RandomForestClassifier, ExtraTreesClassifier
from sklearn.datasets import make_moons, make_classification, make_circles
from sklearn.model_selection import train_test_split
from skgarden import MondrianForestClassifier
sys.path.extend([".", ".."])
from onelearn import AMFClassifier
from experiments import (
get_mesh,
plot_contour_binary_classif,
plot_scatter_binary_classif,
)
logging.basicConfig(
level=logging.INFO, format="%(asctime)s %(message)s", datefmt="%Y-%m-%d %H:%M:%S"
)
np.set_printoptions(precision=2)
n_samples = 1000
random_state = 42
h = 0.01
levels = 20
use_aggregation = True
split_pure = True
n_estimators = 100
step = 1.0
dirichlet = 0.5
norm = plt.Normalize(vmin=0.0, vmax=1.0)
def simulate_data(dataset="moons"):
if dataset == "moons":
X, y = make_moons(n_samples=n_samples, noise=0.2, random_state=random_state)
elif dataset == "circles":
X, y = make_circles(
n_samples=n_samples, noise=0.1, factor=0.5, random_state=random_state
)
elif dataset == "linear":
X, y = make_classification(
n_samples=n_samples,
n_features=2,
n_redundant=0,
n_informative=2,
random_state=random_state,
n_clusters_per_class=1,
flip_y=0.001,
class_sep=2.0,
)
rng = np.random.RandomState(random_state)
X += 2 * rng.uniform(size=X.shape)
else:
X, y = make_moons(n_samples=n_samples, noise=0.2, random_state=random_state)
X = MinMaxScaler().fit_transform(X)
return X, y
datasets = [simulate_data("moons"), simulate_data("circles"), simulate_data("linear")]
n_classifiers = 5
n_datasets = 3
_ = plt.figure(figsize=(2 * (n_classifiers + 1), 2 * n_datasets))
def get_classifiers():
return [
(
"AMF",
AMFClassifier(
n_classes=2,
n_estimators=n_estimators,
random_state=random_state,
use_aggregation=True,
split_pure=True,
),
),
(
"AMF(no agg)",
AMFClassifier(
n_classes=2,
n_estimators=n_estimators,
random_state=random_state,
use_aggregation=False,
split_pure=True,
),
),
(
"MF",
MondrianForestClassifier(
n_estimators=n_estimators, random_state=random_state
),
),
(
"RF",
RandomForestClassifier(
n_estimators=n_estimators, random_state=random_state
),
),
(
"ET",
ExtraTreesClassifier(n_estimators=n_estimators, random_state=random_state),
),
]
i = 1
for ds_cnt, ds in enumerate(datasets):
X, y = ds
xx, yy, X_mesh = get_mesh(X, h=h, padding=0.2)
ax = plt.subplot(n_datasets, n_classifiers + 1, i)
if ds_cnt == 0:
title = "Input data"
else:
title = None
plot_scatter_binary_classif(ax, xx, yy, X, y, s=10, title=title)
i += 1
classifiers = get_classifiers()
for name, clf in classifiers:
ax = plt.subplot(n_datasets, n_classifiers + 1, i)
if hasattr(clf, "clear"):
clf.clear()
if hasattr(clf, "partial_fit"):
clf.partial_fit(X, y)
else:
clf.fit(X, y)
Z = clf.predict_proba(X_mesh)[:, 1].reshape(xx.shape)
if ds_cnt == 0:
plot_contour_binary_classif(
ax, xx, yy, Z, levels=levels, title=name, norm=norm
)
else:
plot_contour_binary_classif(ax, xx, yy, Z, levels=levels, norm=norm)
i += 1
plt.tight_layout()
plt.savefig("decisions.pdf")
logging.info("Saved the decision functions in 'decision.pdf")
Total running time of the script: ( 0 minutes 19.482 seconds)